Xingbo Yang Group
Bioenergetics of Self-Organization

Biological systems display self-organization across scales, from the assembly of subcellular structures to morphogenesis in development. Biological self-organization is characterized by the emergence of order from the interactions among its building blocks, such as molecules and cells. These processes are intrinsically out of equilibrium and driven by energy flows at the scale of its building blocks. It remains a fundamental question to understand how order and structures emerge from interacting building blocks in such processes. While extensive research is being conducted to explore the contribution of gene regulation, signaling and mechanical forces to biological self-organization, much less is known about the contribution of energetics in such processes. My group aims to address fundamental questions in the bioenergetics of self-organization, including:
- What are the energetic costs of fundamental cellular processes, such as cell division, cell motility, ion homeostasis, biosynthesis and information processing?
- How do cellular energy expenditures vary in space and time and how do cells control energetic fluxes spatiotemporally?
- What are the energetic constraints and tradeoffs associated with biological self-organization?
- What are the impacts of energetic variations and perturbations in physiological and pathological conditions?
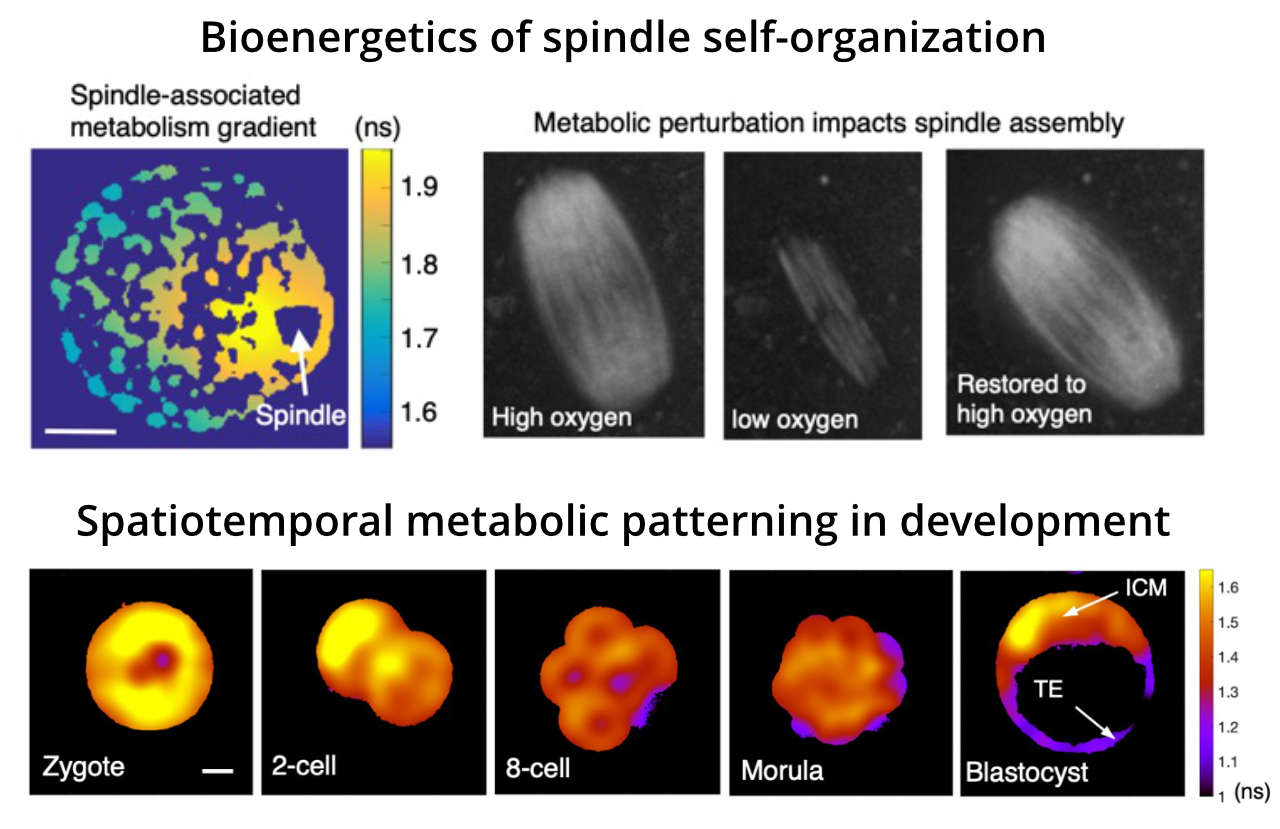
Metabolism supplies free energy for cells. Our group is developing quantitative imaging techniques to measure metabolic activities in living cells, enabling the characterization of spatiotemporal dynamics of metabolic pathways with single-cell and even subcellular resolution. We are combining biophysical modeling with quantitative metabolic imaging to study biological self-organization by exploring the spatiotemporal variation of metabolic activities in cells, tissues and organisms.
Future Projects and Goals
- Combine biophysical modeling with quantitative metabolic imaging to quantify the energetic costs of key cellular processes and to understand how metabolic fluxes are distributed across the cell.
- Understand how perturbations of energy metabolism impact spindle self-organization in meiotic and mitotic cells.
- Characterize the spatiotemporal dynamics of metabolism in development and explore the impact of metabolic variations on cell motility, proliferation and cell fate specification.
Methodological and Technical Expertise
- Quantitative live cell metabolic imaging (e.g. two-photon confocal fluorescence lifetime imaging)
- Biochemical measurements of cell metabolism (e.g. oxygen consumption rate, nutrient uptake rate)
- Retrieval, culturing and microinjection of oocytes and embryos
- Quantitative image analysis
- Computation and mathematical modeling of living systems