Natalie Dye Group
Biophysics of epithelial growth and tumorigenesis

Our group researches the fundamental mechanisms regulating epithelial tissue growth and morphogenesis in development and cancer. Our ignorance on this topic limits efforts to engineer tissues for regenerative medicine, understand pathological tissue morphologies, and explain the evolution of morphological diversity. While many genes have been implicated in regulating tissue morphology, the mechanisms are often unclear because of missing information at the cellular scale – how do genes impact cellular motion? How is cell behavior coordinated in space and time to shape a growing tissue? How does such precise control go wrong when tissues overgrow and metastasize in cancer? Answering these questions requires understanding not just the genetic underpinnings of tissue morphology, but also the mechanical forces influencing cell behavior.
My goal is to elucidate the genetic and physical mechanisms regulating tissue morphology and harness this knowledge for the management of human cancers of epithelial origin. Our approach to this research topic involves bridging the molecular to tissue scales, using quantitative timelapse live imaging, genetic and physical perturbation, and biophysical modeling to uncover the physical principles that organize collective cell behavior. Specifically, my group has two main research directions:
Emergence of tissue organization and shape during development
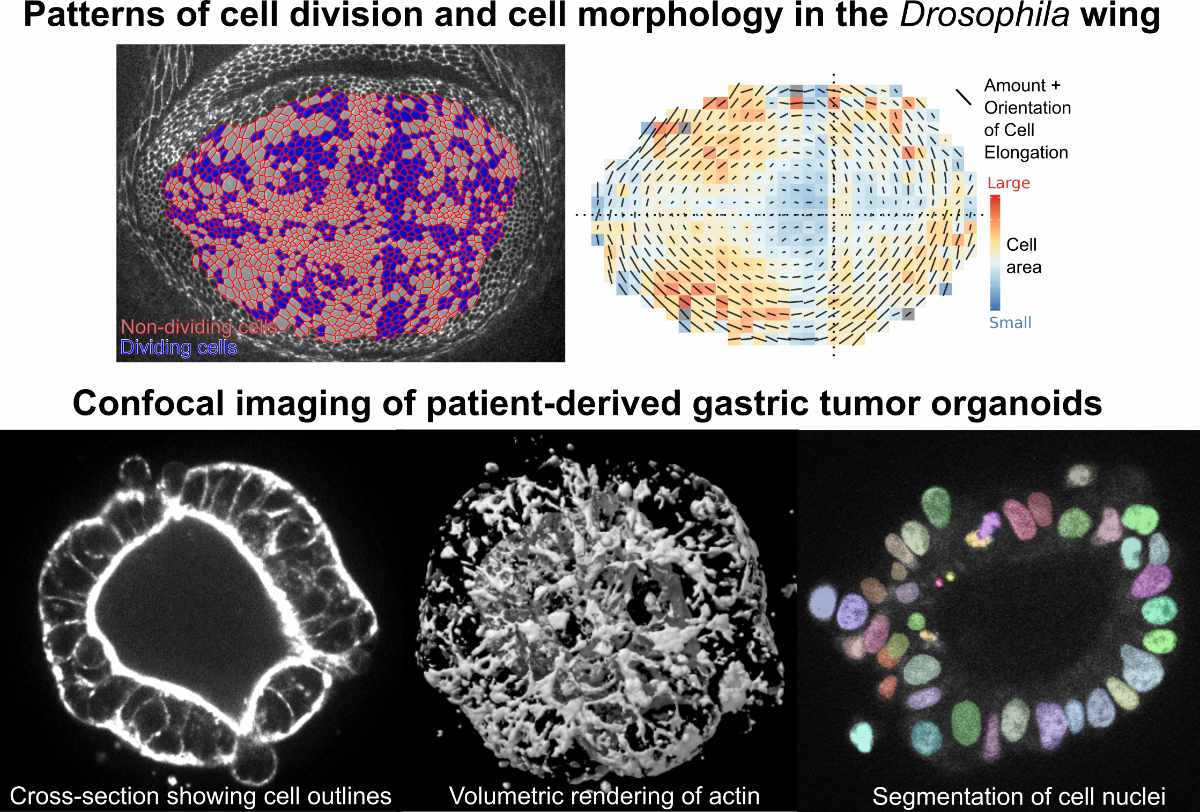
To study the emergence of tissue form during development, our group has been focusing on the model system of the Drosophila wing, due to its genetic tractability and relatively simple, almost 2D geometry. With this system, we have developed methods for (1) long term live imaging of tissue growth and morphogenesis at cellular resolution, (2) quantitatively assessing how cellular behavior deforms the tissue, (3) measuring the mechanical stresses in the tissue, and (4) applying theoretical models to uncover new physical principles. Our systematic analysis of cellular dynamics revealed a potential mechanical constraint guiding anisotropic wing growth. Furthermore, we found a novel type of pattern-formation mechanism, whereby mechanosensitive feedback between stress and polarity can lead to self-organized patterns of cell shape and size. Our work lays the groundwork for investigating how genetic mutations influence tissue morphology and provides a valuable toolset for high resolution analysis of collective cellular behavior. Currently, we are adapting our methods and approach to examine more complex 3D tissue architectures, using both Drosophila and human stem cell systems.
A mesoscale, biophysical approach to precision oncology
We are now motivated to apply the concepts, approach, and methodology that we have gained from developmental biology to the realm of human disease. Specifically, we are developing a novel, biophysical approach to precision oncology, with the ultimate aim of improving our ability to individualize cancer treatments. Toward this end, we are using recently developed methods for propagating human stem cells in 3D ex vivo culture as “organoids” – small cell collectives that resemble the host tissue architecture and gene expression. We aim to analyze the dynamics and mechanics of patient-derived tumor organoids to gain a new type of mechanistic insight into tumor progression and design cellular-level metrics for predicting disease outcome and drug response. Such an analysis will provide a missing link between known variation at the molecular scale and the complex, large scale phenotype of cancer growth and metastasis.
Future Projects and Goals
We aim to: (1) uncover mechanisms that transform epithelial sheets into complex tissue shapes during development, and (2) develop novel methods for analyzing individual variation in epithelial tumorigenesis and metastasis in human cancer patients. With funding provided by the Mildred-Scheel-Nachwuchszentrum (MSNZ – Mildred Scheel Early Career Center), we are specifically expanding this second area of research. Those with expertise in physics, programming, and/or microscopy who are motivated to transition to cancer biology and biomedicine are especially encouraged to apply. The goals of this project include:
- Connect organoid architecture and dynamics with cancer patient outcome.
- Determine the biophysical mechanisms underlying variability in organoid morphology.
- Design novel cellular-level metrics for predicting cancer severity and drug response.
Methodological and Technical Expertise
- Confocal and light sheet microscopy
- Quantitative image analysis of tissue architecture and dynamics
- Human gastrointestinal organoids
- Biophysical modeling
- Drosophila genetic