Mareike Albert Group
Gene regulatory mechanisms of neocortex evolution

© CRTD
The neocortex is considered to be the seat of higher cognitive functions in humans. During its evolution, most notably in humans, the neocortex has undergone considerable expansion, which is reflected by an increase in the number of neurons. Neocortical neurons are generated during development by neural stem and progenitor cells. Epigenetic mechanisms play a pivotal role in orchestrating the behaviour of stem cells during development. We are interested in the mechanisms that regulate gene expression in neural stem cells, which have implications for our understanding of neocortex development and evolution, neural stem cell regulation and neurodevelopmental disorders.
Epigenetic regulation of human neocortex development
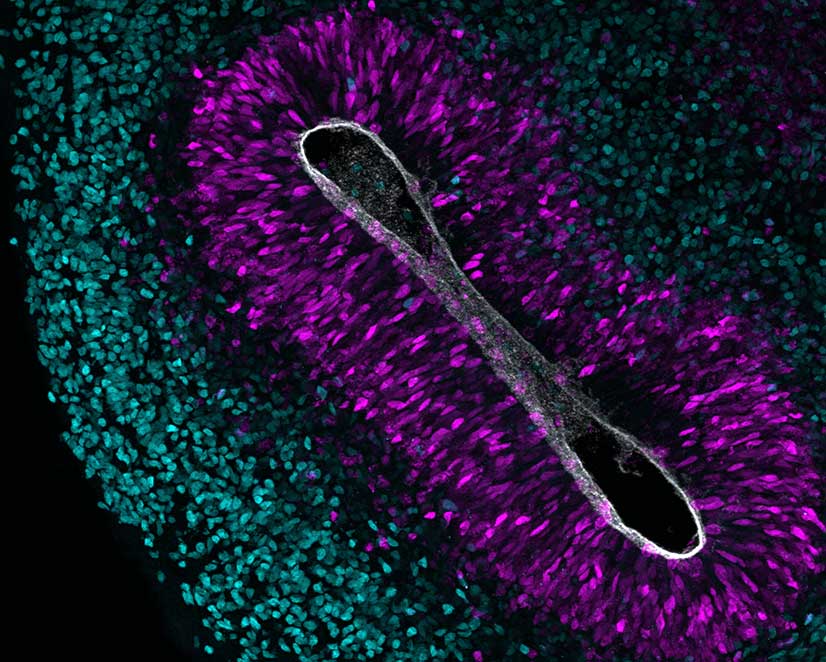
During the development of the neocortex, stem and progenitor cells initially proliferate, then sequentially give rise to neurons destined to different cortical layers and finally switch to gliogenesis resulting in the generation of astrocytes and oligodendrocytes. Precise spatial and temporal regulation of neural progenitor differentiation is key for the proper formation of the complex structure of the neocortex. An impairment of the development of the brain, as observed in patients with neurodevelopmental disorders, is frequently linked to compromised intellectual abilities.
Chromatin modifiers of the Polycomb group of proteins are known to regulate important aspects of neocortex development (Albert and Huttner, 2018; Hoffmann and Albert, 2021). By epigenome profiling in specific neural cell populations, we have shown that histone methylation is highly dynamic during neocortex development (Albert et al., 2017). Moreover, by CRISPR/Cas9-based epigenome editing, we could show that altering histone methylation at a selected promoter is sufficient to reduce the expression of a key neural transcription factor resulting in altered neural progenitor abundance (Albert et al., 2017). We are currently focusing on the development of the human neocortex and on neurodevelopmental disorders that are caused by mutations in epigenetic players (Bölicke and Albert, 2022).
Evolutionary expansion of the neocortex
Interspecies differences in both neuron number and neocortex size are thought to arise from differences in the proliferative capacity and the abundance of neural progenitor subtypes. Specifically, basal progenitors, such as basal radial glia, are particularly abundant in mammals with a large, folded neocortex and characterized by extensive self-renewal and proliferative capacities. Transcriptome studies in different mammalian species have uncovered genes that are expressed in human basal radial glia, among which are the human-specific genes ARHGAP11B and NOTCH2NL that may have contributed to neocortex expansion during evolution (Florio et al., 2015; Florio et al., 2018). Moreover, we found that the growth factor EPIREGULIN is differentially expressed between species and may have contributed to neocortex expansion in primates by providing a tuneable pro-proliferative signal to basal progenitor cells in the subventricular zone progenitor niche (Cubillos et al, 2024).
The gene regulatory mechanisms that orchestrate specific gene expression programs from the genomic DNA in different neural progenitor cell types in different species remain poorly understood. Gene transcription is initiated at promoter sequences immediately upstream of a gene, but differential gene expression in specific cell types is controlled by often distally located regulatory elements. This complex regulatory landscape allows cells to respond to developmental and environmental stimuli. We are studying gene regulation using in vivo models and in vitro human iPCS-derived 3D brain organoids (Schütze et al, 2022). Our work aims to provide new insights into gene regulatory mechanisms with implications for neocortex evolution as well as neuropathologies.
Future Projects and Goals
The aims of our research are
- to elucidate the epigenetic regulation of human neocortex development
- to further develop models of neurodevelopmental disorders caused by mutations in epigenetic factors
- to understand how epigenetic mis-regulation contributes to neuropathologies and whether epigenetic changes and phenotypes can be reversed
- to identify and characterize gene regulatory regions that may have contributed to human brain evolution
Methodological and Technical Expertise
- In vivo and in vitro models of neocortex development
- Generation of human cortical organoids from iPSC
- Genome and epigenome editing using CRISPR/Cas9
- Genomics (RNA-seq, ChIP-seq, ATAC-seq, CUT&Tag)
- Isolation of neural stem and progenitor cells (FACS)
- Immunohistochemistry and microscopy
- Single cell epigenome analysis (Epi-CyTOF)