Lutz Brusch Group
Spatio-temporal pattern formation in cells and tissues

Feedback regulation is ubiquitous in nature, starting already at the molecular scale and extending all the way up, often linking different spatial scales. Biological systems utilize feedback regulation to let structures and functions self-organize. Such self-organization often defies our linear-pathway-trained intuition. Mathematical modeling, analysis and computer simulation closely iterated with quantitative experiments can therefore be a fruitful strategy for studying complex regulatory systems.
We have previously applied this theoretical approach in collaboration with experimentalists and found that feedback loops of effector recruitment and activity can couple different Rab GTPases on intracellular organelles in such a way that multistability self-organizes (del Conte-Zerial et al., 2008). Therewith, membrane identity is robustly maintained despite of noise and perturbations but can also abruptly switch to a successive identity upon a super-threshold signal. This abrupt switch, as observed for instance in early to late endosome conversion, represents a novel type, termed cut-out switch, which differs from the canonical toggle switch by superposition of negative feedback upon the positive feedback loop and correspondingly novel switch behavior. Such theoretical insight into biological switches was also applied in the design and analysis of biosynthetic computing circuits (Hayat et al., 2006). Currently, self-organizing multistability and switches between different functional states are further explored for pancreas cell differentiation with implications for cell reprogramming in regenerative medicine (Zhou et al., 2011).
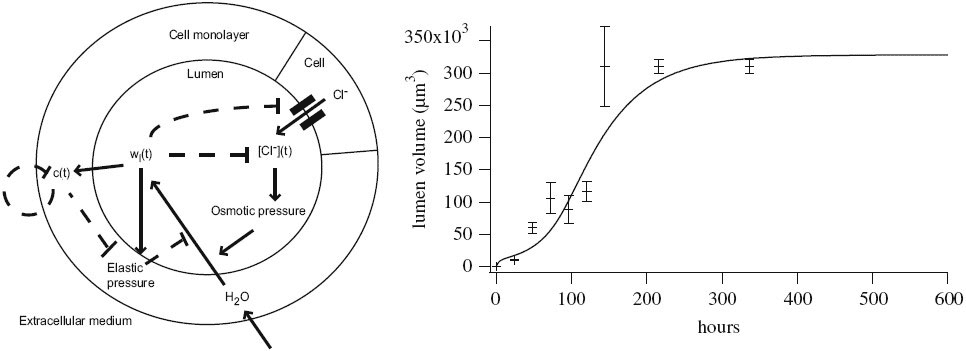
More complex spatio-temporal behavior emerges when regulatory networks couple spatially distributed components. Along this line, a mechanistic model and analysis of ErbB signal transduction and gene regulation revealed that differentiation versus proliferation decisions of cells upon growth factor stimulation arise from spatially distributed signal processing (Nakakuki et al., 2010). We are currently also exploring how feedback regulation and spatio-temporal patterning control tissue morphogenesis and growth (Gin et al., 2010).
Throughout these projects, the mathematical models are inherently nonlinear and only fully disclose their secrets through advanced methods including dynamical systems theory and bifurcation analysis. We continuously develop such analysis methods further, in particular we’re the team behind the user-friendly modeling and simulation framework Morpheus.
Future Projects and Goals
This theory group, in close collaborations with experimentalists, will further extend and apply systems biology approaches to uncover the regulatory mechanisms that underlie spatio-temporal patterning processes in cells and tissues.
Open questions that inspire our work are
- Which capabilities does the eukaryotic cell gain from the closed regulatory loop between signal transduction and compartmentalization/endocytosis?
- How is hepatocyte polarity established and remodeled in the regenerating liver?
- What kind of biochemical and/or biophysical signals does a growing, and moreover a regenerating, tissue feed back from the tissue/organ scale down to a single proliferating cell such that a robust organ size emerges?
Methodological and Technical Expertise
- Mathematical modeling of processes in cells and tissues using Morpheus
- Model analysis using dynamical systems theory, especially bifurcation analysis
- Computer simulation and high performance computing
- Data analysis and parameter estimation