Henrik Bringmann Group
How to regenerate during sleep

The molecular mechanisms underlying the vital functions of sleep in promoting health are not understood. This is surprising since sleep is essential and sleep disorders are highly prevalent in industrialized societies, posing a massive unsolved medical and economic challenge. The goal of our lab is to solve how sleep becomes regenerative at the molecular level and how it supports development, regeneration, immunity, cellular health and a long life. To solve key sleep functions, we are generating and analyzing genetic sleeplessness in animal models.
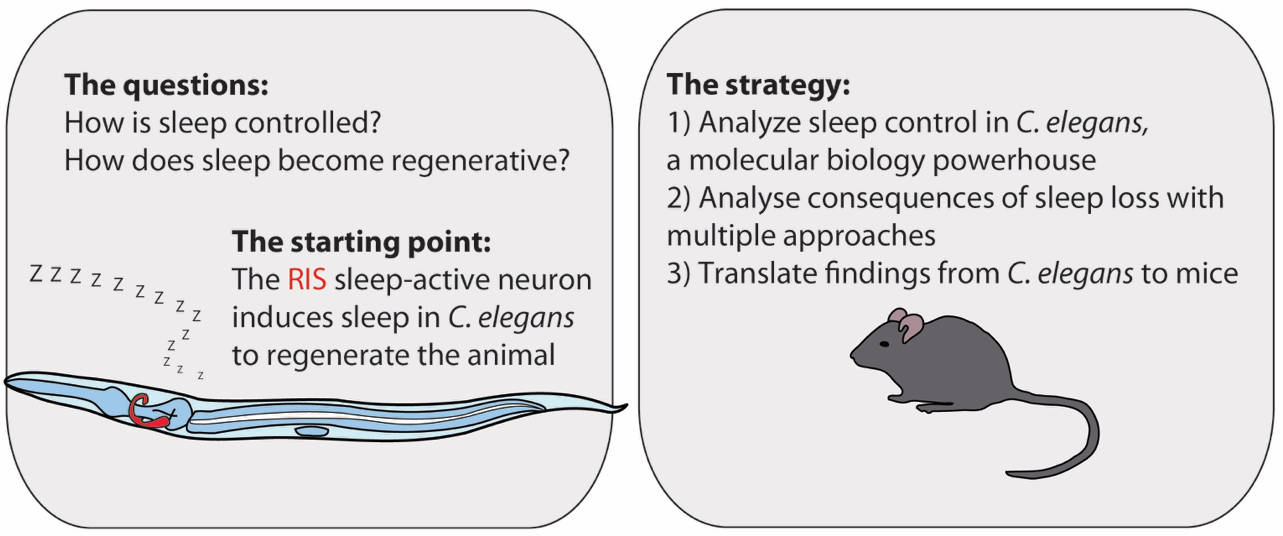
All animals sleep, at least those that have a nervous system, suggesting that this physiological state emerged early in the course of evolution and is conserved. To understand sleep, we have to understand how it is controlled and how it exerts its functions at the molecular and cellular level. To solve these questions, our lab has embarked on a long-term research plan to first figure out how sleep is controlled in C. elegans, as the worm is the simplest model animal that sleeps and that is accessible to molecular studies. The molecular knowledge about sleep control is now being used to genetically deprive C. elegans of sleep and thus to understand the molecular mechanisms underlying its cell biological and biochemical functions. Finally, we are translating our findings to mouse models.
Sleep deprivation (SD) by sensory stimulation is the classic method used to study sleep functions, but this approach can also cause stress and unspecific side effects, thus hampering a molecular dissection of molecular sleep functions. A novel alternative strategy to remove sleep is to ablate it by using mutation. Sleep-active neurons induce sleep and impairing them or their regulators can lead to specific sleep loss. However, complete and specific genetic SD is not yet possible in most models. Our lab found that C. elegans possesses a single key sleep-active neuron. Impairing this neuron genetically or optogenetically leads to virtually complete and highly specific sleep loss, generating a unique model for genetic SD. Genetic SD can blunt survival and increases the progression of aging phenotypes, but the underlying mechanisms are not understood. We are analyzing sleepless C. elegans mutants with a combination of genetic screening, omics, and cell biological analyses, which will result in a molecular and mechanistic understanding of sleep functions.
We are using the molecular sleep information from C. elegans as a roadmap to understand mammalian sleep. For this, we are knocking down homologs of C. elegans sleep genes in mice. We could show that AP-2 transcription factors evolved first as sleep promoters in invertebrates and then diversified to bidirectionally control sleep in mammals. We use genetic sleep deprivation mouse models to study mammalian sleep functions. Ultimately, our translational research will lead to an understanding of human sleep and will help develop treatments for human insomnia and to develop regenerative therapies that build on the molecular benefits exerted by sleep.
Future Projects and Goals
- We will identify conserved genes, neuronal circuits, and cellular mechanisms for the control of sleep, with a particular emphasis on the functioning of sleep-active neurons. For this goal, we are using the power of C. elegans genetics and then translate our findings to mice.
- We will solve the molecular functions of sleep in cellular health, immunity, development, regeneration, and aging by analyzing the consequences of genetic sleep loss in our animal models.
- Based on our work regarding the regulation and functions of sleep in model organisms, we will devise strategies for developing therapies for human sleep disorders and regenerative medicine.
Methodological and Technical Expertise
- Calcium imaging
- Optogenetics
- C. elegans genetics
- Automated in-vivo imaging
- Image processing