Felix Müller-Planitz Group
Epigenetics and Chromatin: How chromatin forms and how it affects cellular functions

© Jan Greune
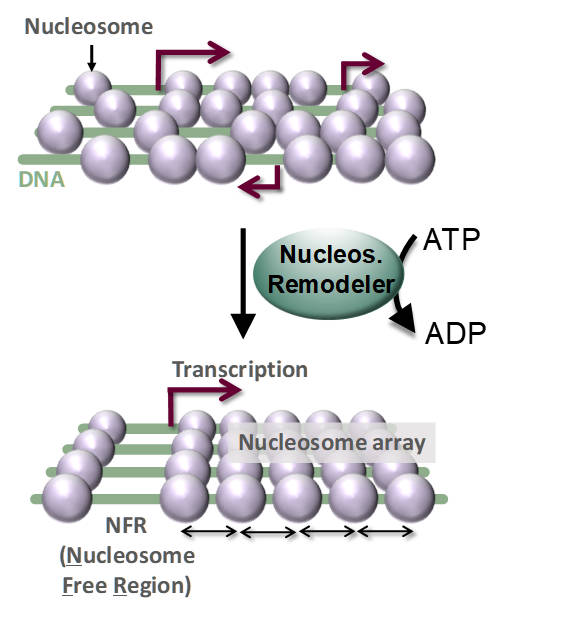
The Mueller-Planitz lab studies the core components of chromatin – the nucleosomes and the machinery that places them in the genome. Nucleosomes are crucial to human health. Aging, for instance, disrupts the nucleosome landscape, destabilizing the genome, and mutations in nucleosomes are drivers of cancers. Nucleosomes serve both as barriers that restrict access to the genome and as a medium to accumulate epigenetic marks. Correspondingly, the locations of nucleosomes in the genome are precisely controlled by so called nucleosome remodeling complexes. Remodelers move, assemble, or eject nucleosomes in an ATP-dependent fashion. Some also even the spacing between nucleosomes, setting a characteristic nucleosome-to-nucleosome distance. These “spacing remodelers” thereby generate arrays of nucleosomes with a surprising regularity, and these arrays are conserved throughout eukaryotes. Their function however remains elusive.
The overarching aim of the Mueller-Planitz lab is to elucidate the biogenesis of the nucleosome landscape and dissect its biological function under physiological and pathological conditions. To achieve this goal, his lab bridges methodologies of molecular biology, genetics, genomics, biophysics, structural biology, and enzymology. They develop cutting-edge technology to visualize individual nucleosome patterns in single cells, to deduce systems-level properties of tens of thousands of nucleosomes in cells, and to dissect the mechanism of nucleosome remodeling genome-wide in vivo and in vitro.
Future Projects and Goals
- Changes in chromatin during natural and premature aging
- Transcription through chromatin
- Conformational dynamics of nucleosome remodelers
- Chromatin remodeling in phase-separated chromatin
- Biogenesis and maintenance of heterochromatin
Methodological and Technical Expertise
- Next Generation and Third Generation Sequencing techniques
- Third generation single molecule sequencing such as Nanopore sequencing
- FRET, single-molecule FRET
- FRAP, FLIM