Anna Poetsch Group
Biomedical Genomics

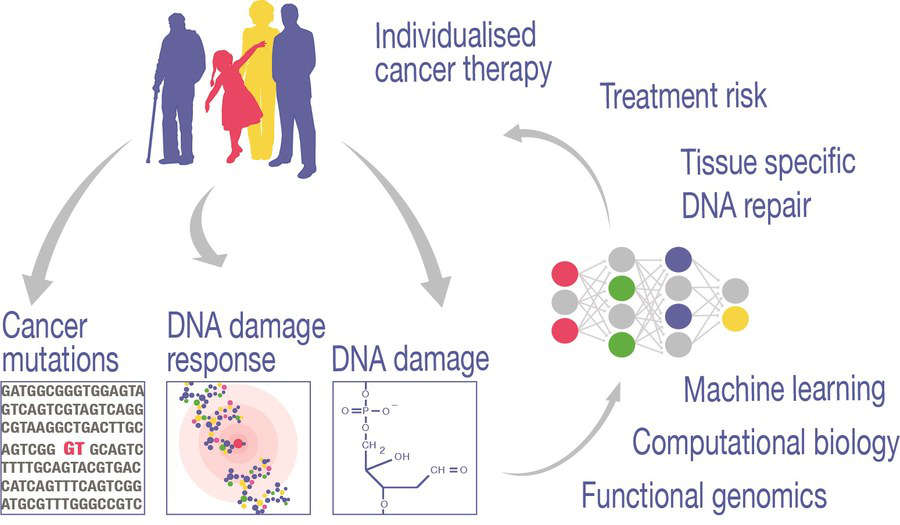
We study the genomics of DNA damage, repair, and mutagenesis. Using a combination of computational biology, machine learning, clinical and experimental data from collaborating labs, we try to understand the underlying mechanisms despite their complexity, and look for routes to bring this knowledge into clinical use.
We develop novel techniques to measure oxidative DNA damage genome-wide and establish the associated data analysis strategies. Thus, we found an unappreciated mechanism that leads to lower damage rates in coding sequence, yet accumulation of damage in specific repetitive DNA. This is partially reflected in the related mutation profiles of cancer genomes. The mechanisms that lead to the specific distribution are very poorly understood. Are we maybe even mistaking oxidative DNA "damage" as something entirely unwanted? Could there be a second sign to the coin, a function as a stress sensor, a role as an epigenetic mark?
To understand these processes, we are also making use of the large amounts of data available to us in the form of sequenced cancer genomes. DNA damage processes left their mark in the form of mutations at specific locations in the genome. While these are noisy data, the snapshot from multiple mutagenic processes in each sample, the information of their history is contained in the patterns. We use deep learning to use such patterns and extract the biology of mutagenesis.
Major focus areas
- The genomics of DNA damage, repair, and DNA damage response
This area combines several projects that we pursue together with collaborators on mechanistic questions in the DNA damage response. Combining biochemical data with functional genomics approaches, these highly interdisciplinary projects keep us up to speed with the cutting edge mechanistic questions that currently occupy the field. - Learning genome specificity of mutagenesis
We are using cascade deep learning models that combine classification and regression tasks to investigate mutagenic mechanisms and their interactions. The models are trained on thousands of whole genome sequencing datasets to model the specificity of mutagenic mechanisms. We use these models as an in silico experimental system to answer hypotheses on mutation specificity by simulating the conditions we are interested in.
In a complementary approach we predict per-nucleotide mutation rates with recurrent neural networks (LSTMs) for individual patients. - Learning the language of instability through large language models on genomic data
The genetic code is frequently viewed from the perspective of how proteins are encoded in the DNA. Yet, also the probability to mutate, the stability of the double helix, the ability to form non-B-DNA, and the location of repetitive sequence is to some extent also encoded in the DNA, we just don't understand it.
We are trying to tackle this ignorance with the help of large language models that also in linguistics now give unprecedented possibilities to quantitatively describe grammar, semantics and other language rules. Why not using DNA as a text and do the same? In addition to a basic understanding of how the language of DNA works beyond protein coding, using language models for targeted machine learning tasks show much increased performance and can be used for example to predict mutation rate distribution with unprecedented accuracy and precision of CRISPR based genome editing.
Specific current areas of interest
- Learning mutagenesis
- Oxidative DNA “damage”
- Learning the language of the human genome
- DNA damage response
- Machine learning in personalised medicine
Future Projects and Goals
The goal of the group is to understand the genomics of DNA damage, repair, and mutagenesis. This is a very interdisciplinary task, as it requires an understanding of chemistry, molecular biology, how the genome works, and at the same time the methodology is quantitative, computational and includes cutting edge methods in machine learning and natural language processing. We will do all this with keeping a close link to the clinical oncologists and interdisciplinary consortia, because we ultimately want to use our gained knowledge for application in the clinic.
Bringing these ostensibly separate ways of thinking together will be part of any future project of the group, irrespective of whether it will be part of looking into mechanisms of the DNA damage response, looking at mutations in cancer, or deep into deep learning of how the genome works.
Methodological and Technical Expertise
- Functional genomics (multi-omics in bulk and single cell; wet and dry)
- X-Seq data analysis, including (sc)RNA-Seq, ChIP-Seq, ATAC-Seq, DRIP-Seq, iCLIP, CAGE-Seq, END-Seq, HiC, …
- Oxidative DNA damage genome-wide measurements and data analysis
- Cancer genomics
- Genome editing data analysis
- Machine Learning
- Clinical Outcome Predictions (with machine learning)
- Deep Learning
- Natural Language Processing (on genomes)