Michael Schlierf Group
Conformational changes in biomolecules and enzymes

© TUD CMCB, Magdalena Gonciarz
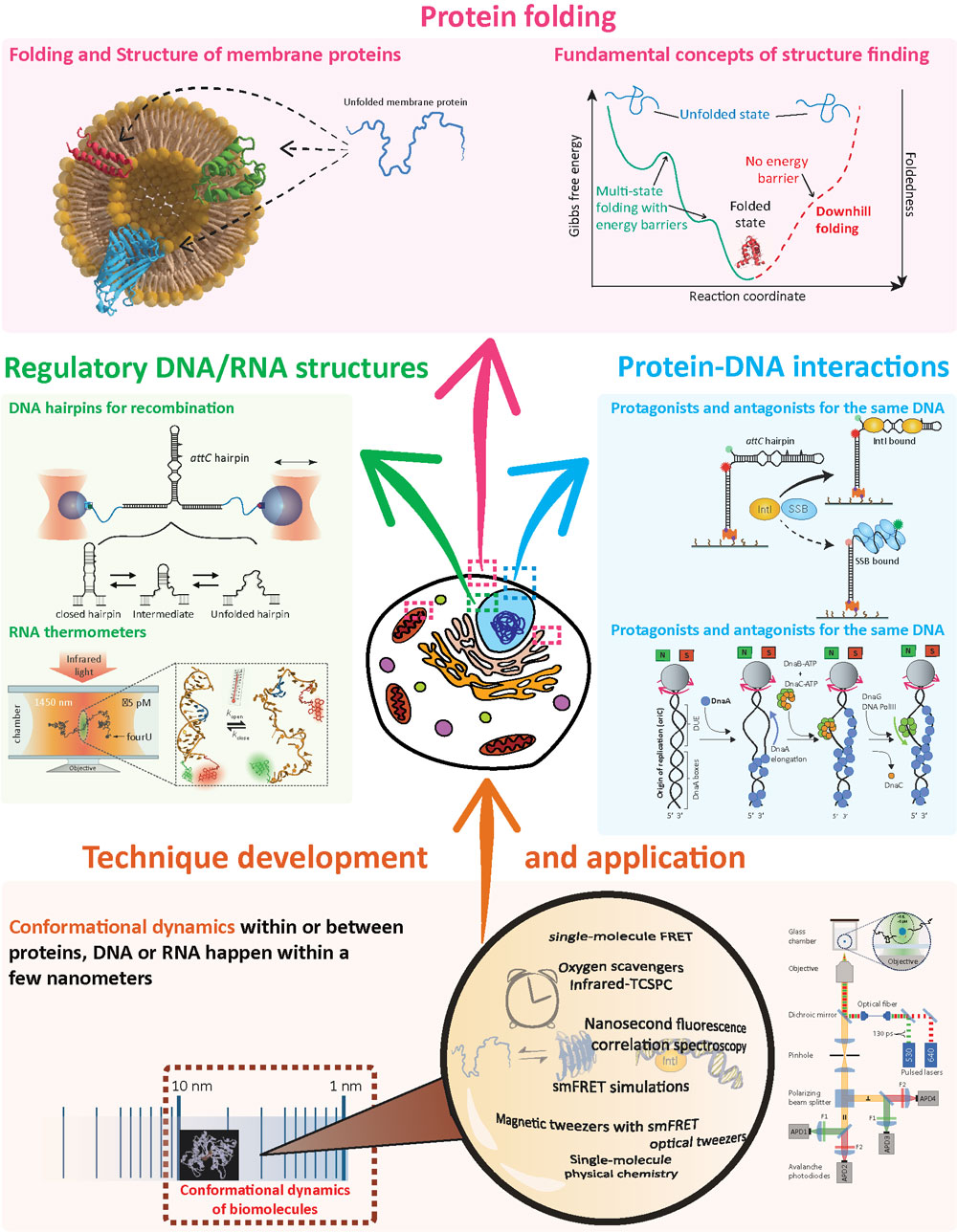
The cells that keep our bodies working are like factories run by millions of tiny machines made of DNA, RNA, proteins and lipids. They store the genetic information of our organism, allow our cells to grow and replicate, defend our system against infection and transport nutrients. They achieve this by being dynamic and changing their 3D-structure over time and as a response to stimuli. In particular, our lab studies membrane protein folding, protein-DNA interactions during DNA replication and recombination and regulatory DNA and RNA structures.
Membrane proteins are key players in the functioning of cells. They are located at the interface between the inside and outside of cells and compartments. Precise folding and occupation of the accurate tertiary structure within the membrane is crucial for their correct functioning and can be considerably influenced by the surrounding lipid composition. Already single point mutations in the amino acid chain can lead to severe structural changes and diseases, like cystic fibrosis. Their hydrophobic nature and the often complex lipidic environment creates challenges to study folding and insertion processes. We developed methods to study folding and unfolding of membrane proteins in lipidic environments using single-molecule FRET. We aim to dissect fundamental principles from the highly complex process of structure formation and transition into the anisotropic environment of lipid bilayers.
Protein-DNA interactions are the key for cell regulation, including cell division and adaptation. There are different levels of specificity for these interactions ranging from unspecific single-stranded DNA binding to specific secondary structure recognition. The protein DnaA, along with other molecules, is essential for opening the DNA double helix during DNA replication. We reconstitute in a bottom-up single-molecule approach this process to develop a picture of which key interactions make our genome getting duplicated. A second example is the bacterial Integron system, in which a DNA secondary structure can trigger and regulate DNA recombination during adaptation processes within bacteria, including the transfer of antibiotic resistances. Often these DNA structures are recognized by specific recombinases, which face and master the challenge with antagonists.
DNA and RNA are known to form secondary structures, if self-complementary sequences are present. RNA secondary structures have evolved as expression controls (riboswitches) and DNA secondary structures are used as recombination sites (Integron). Here, we study the structure and conformational kinetics of RNA and DNA hairpin structures. One example is the RNA molecule fourU, a heat-shock response in bacterial cells. We study the dynamics and responses of this simple, yet, efficient RNA hairpin structure.
In order to study conformational changes within and between biomolecules, we develop and apply modern instruments and techniques based on fluorescence and force spectroscopy to understand how these nano-bio-machines change their structure during a given biological process: we are able to observe and analyze one molecule at a time to build a highly time and space-resolved picture of their dynamic interaction.
Future Projects and Goals
In order to study conformational changes within and between biomolecules, we develop and apply modern instruments and techniques based on fluorescence and force spectroscopy to understand how these nano-bio-machines change their structure during a given biological process: we are able to observe and analyze one molecule at a time to build a highly time and space-resolved picture of their dynamic interaction.
Main research fields include:
- membrane protein folding
- protein-DNA interactions
- regulatory DNA and RNA conformations
Methodological and Technical Expertise
- molecular bioengineering
- single-molecule FRET
- TCSPC
- magnetic tweezers
- optical tweezers